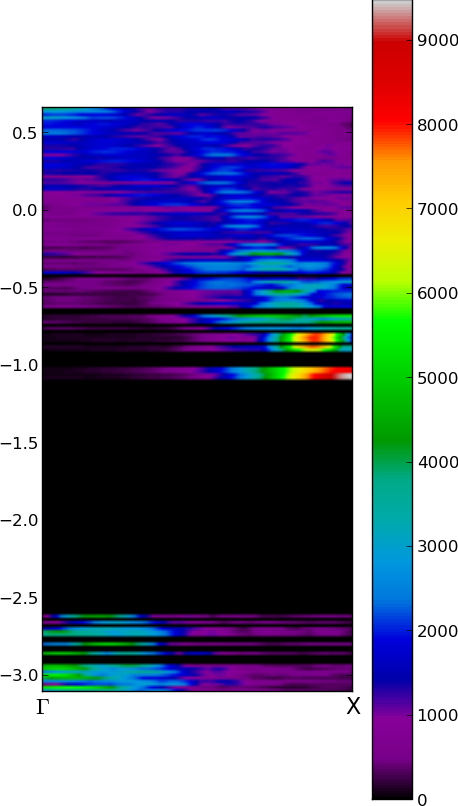
band_structure_of_nanocrystals
This is an old revision of the document!
Table of Contents
Band structure of nanocrystals
There is a possibility to get view of fuzzy band structure by Fourier transform of eigenstates of nanocrystal (i.e. cluster without periodic boundary condition). See Hapala et.al,Phys.Rev.B87,195420.
Availability
this is implemented only in Prokop's personal versions of Fireball located in
/data/home/hapala/Fireball_dev/src_1.0-BSfinal /data/home/hapala/Fireball_dev/progs_Jellium_mod
3D Fourier fransform of Molecular Orbitals
fireball.in
&OPTION basisfile = answer.bas nstepf = 1 icluster = 1 ifixcharge = 1 dt = 0.5 &END &OUTPUT iwrtewf = 1 &END &MESH iewform = 5 npbands = 5 pbands = 159,160,161,162,163 &END
1D cuts of Fuzzy Band Structure
fireball.in
&OPTION basisfile = answer.bas nstepf = 1 icluster = 1 ifixcharge = 1 dt = 0.5 &END &OUTPUT iwrtewf = 1 &END &MESH iewform = 6 &END
kscan.optional
1 # byEnerg -8.0000 1.0000 # Emin Emax 5.50 # alat 48 100 # nlines nkpoints 2.0 0.0 0.0 1.0 1.0 0.0 2.0 0.0 0.0 1.0 -1.0 0.0 2.0 0.0 0.0 1.0 0.0 1.0 2.0 0.0 0.0 1.0 0.0 -1.0 0.0 2.0 0.0 1.0 1.0 0.0 0.0 2.0 0.0 -1.0 1.0 0.0 0.0 2.0 0.0 0.0 1.0 1.0 0.0 2.0 0.0 0.0 1.0 -1.0 0.0 0.0 2.0 -1.0 0.0 1.0 0.0 0.0 2.0 1.0 0.0 1.0 0.0 0.0 2.0 0.0 1.0 1.0 0.0 0.0 2.0 0.0 -1.0 1.0 -2.0 0.0 0.0 -1.0 1.0 0.0 -2.0 0.0 0.0 -1.0 -1.0 0.0 -2.0 0.0 0.0 -1.0 0.0 1.0 -2.0 0.0 0.0 -1.0 0.0 -1.0 0.0 -2.0 0.0 1.0 -1.0 0.0 0.0 -2.0 0.0 -1.0 -1.0 0.0 0.0 -2.0 0.0 0.0 -1.0 1.0 0.0 -2.0 0.0 0.0 -1.0 -1.0 0.0 0.0 -2.0 1.0 0.0 -1.0 0.0 0.0 -2.0 -1.0 0.0 -1.0 0.0 0.0 -2.0 0.0 1.0 -1.0 0.0 0.0 -2.0 0.0 -1.0 -1.0 1.0 1.0 1.0 0.0 1.0 1.0 1.0 1.0 1.0 1.0 0.0 1.0 1.0 1.0 1.0 1.0 1.0 0.0 -1.0 1.0 1.0 0.0 1.0 1.0 -1.0 1.0 1.0 -1.0 0.0 1.0 -1.0 1.0 1.0 -1.0 1.0 0.0 1.0 -1.0 1.0 0.0 -1.0 1.0 1.0 -1.0 1.0 1.0 0.0 1.0 1.0 -1.0 1.0 1.0 -1.0 0.0 -1.0 -1.0 1.0 0.0 -1.0 1.0 -1.0 -1.0 1.0 -1.0 0.0 1.0 -1.0 -1.0 1.0 -1.0 -1.0 0.0 1.0 1.0 -1.0 0.0 1.0 -1.0 1.0 1.0 -1.0 1.0 0.0 -1.0 1.0 1.0 -1.0 1.0 1.0 0.0 -1.0 1.0 -1.0 0.0 1.0 -1.0 -1.0 1.0 -1.0 -1.0 0.0 -1.0 -1.0 1.0 -1.0 -1.0 1.0 0.0 1.0 -1.0 -1.0 0.0 -1.0 -1.0 1.0 -1.0 -1.0 1.0 0.0 -1.0 1.0 -1.0 -1.0 1.0 -1.0 0.0 -1.0 -1.0 -1.0 0.0 -1.0 -1.0 -1.0 -1.0 -1.0 -1.0 0.0 -1.0 -1.0 -1.0 -1.0 -1.0 -1.0 0.0
klines_XXXXX.dat
1 391.30476603 391.30476603 391.30476603 391.30476603 287.45162513 ...
2 391.20959758 391.21921233 390.08793464 391.61800721 295.99428664 ...
3 389.17880205 389.77130848 387.81208053 388.10778349 306.53352273 ...
klines_TOT.dat
Ei rho(k1) rho(k2) rho(k3) rho(k4) rho(k5) ... -7.99755 413.91048549 428.76133304 440.87314746 450.14882297 456.52837031 ... -7.99140 394.18797633 403.02567283 409.86012222 414.38007661 416.29758353 ... -7.97472 418.51477832 433.83793597 448.50901654 462.18377663 474.53904963 ... -7.96729 262.72449682 269.69434762 274.26597685 275.89514172 285.23659456 ...
plotting results
#!/usr/bin/python
from pylab import *
import numpy
dE = 0.02 # [ eV ]choose width of energy-bins for plotting
F = transpose(genfromtxt( 'klines_TOT.dat' ))
Es = F[0]
Ps = transpose(F[1:])
Emin =Es.min(); Emax =Es.max()
Pgrid = zeros( ( int( (Emax-Emin)/dE )+1 , shape(Ps)[1] ) )
n = len(Es)
for i in range(n):
iE = int((Es[i]-Emin)/dE)
#Pgrid[iE] += Ps[i] # overlaping (=degenerated) states are summed up
Pgrid[iE] = numpy.vstack([Pgrid[iE],Ps[i] ]).max(axis=0) # for overlaping (=degenerated) states is taken maximum
# choose color map reference here http://wiki.scipy.org/Cookbook/Matplotlib/Show_colormaps
cmap='spectral'
figure( figsize=(5,10) )
extent = ( 0,2, Emin, Emax )
imshow( Pgrid, origin='image', extent=extent, cmap = cmap )
xticks([0,2] , ['$\Gamma$','X'], fontsize=16) # x-axis ticks Gamma and X vector
colorbar()
savefig('FuzzyBand.png', bbox_inches='tight' )
show()
band_structure_of_nanocrystals.1416308052.txt.gz · Last modified: 2014/11/18 11:54 (external edit)