Lattice parameter
In this section, we explain how to optimize the lattice parameter of Ci bulk fcc structure. The fcc structure is characterized by single distinct atom per unit cell with relative coordinates (0 ,0 ,0) . The lattice vector is defined in the relative coordinates alat*(1/4, 1/4, 1/4).
Detail description of the diamond structure can be found elsewhere.
There is an elegant way to rescale atomic coordinates, lattice vector and k-points by only one parameter rescal defined in the section &OPTIONS.
Our input atomic coordinates written in Cu_bulk.bas look like:
1 29 0.00000 0.00000 0.00000
The lattice vector is defined in Cu_bulk.lvs file:
0.70711 0.00000 0.00000 0.35355 0.61237 0.00000 0.35355 0.20412 0.57735
The k-points 8x8x8 Monkhorst-Pack grid can be by Oh symmetry reduced into 128 k-points. They are in Cu_bulk.kpts file:
128 -1.07541645 -0.62089056 -0.10467514 0.00781250 -1.07541645 -0.62089056 -0.07476795 0.00781250 -1.07541645 -0.62089056 -0.04486077 0.00781250 -1.07541645 -0.62089056 -0.01495359 0.00781250 -1.07541645 -0.26609597 -0.10467514 0.00781250 -1.07541645 -0.26609597 -0.07476795 0.00781250 -1.07541645 -0.26609597 -0.04486077 0.00781250 -1.07541645 -0.26609597 -0.01495359 0.00781250 -1.07541645 0.08869865 -0.10467514 0.00781250 -1.07541645 0.08869865 -0.07476795 0.00781250 -1.07541645 0.08869865 -0.04486077 0.00781250 -1.07541645 0.08869865 -0.01495359 0.00781250 -1.07541645 0.44349325 -0.10467514 0.00781250 -1.07541645 0.44349325 -0.07476795 0.00781250 -1.07541645 0.44349325 -0.04486077 0.00781250 -1.07541645 0.44349325 -0.01495359 0.00781250 -1.07541645 0.79828787 -0.10467514 0.00781250 -1.07541645 0.79828787 -0.07476795 0.00781250 -1.07541645 0.79828787 -0.04486077 0.00781250 -1.07541645 0.79828787 -0.01495359 0.00781250 1.38267840 -0.26609593 -0.10467514 0.00781250 1.38267840 -0.26609593 -0.07476795 0.00781250 1.38267840 -0.26609593 -0.04486077 0.00781250 1.38267840 -0.26609593 -0.01495359 0.00781250 1.38267840 0.08869869 -0.10467514 0.00781250 1.38267840 0.08869869 -0.07476795 0.00781250 1.38267840 0.08869869 -0.04486077 0.00781250 1.38267840 0.08869869 -0.01495359 0.00781250 -1.07541645 -0.97568511 -0.10467514 0.00781250 -1.07541645 -0.97568511 -0.07476795 0.00781250 -1.07541645 -0.97568511 -0.04486077 0.00781250 -1.07541645 -0.97568511 -0.01495359 0.00781250 -0.76815462 -0.79828787 -0.10467514 0.00781250 -0.76815462 -0.79828787 -0.07476795 0.00781250 -0.76815462 -0.79828787 -0.04486077 0.00781250 -0.76815462 -0.79828787 -0.01495359 0.00781250 -0.76815462 -0.44349325 -0.10467514 0.00781250 -0.76815462 -0.44349325 -0.07476795 0.00781250 -0.76815462 -0.44349325 -0.04486077 0.00781250 -0.76815462 -0.44349325 -0.01495359 0.00781250 -0.76815462 -0.08869865 -0.10467514 0.00781250 -0.76815462 -0.08869865 -0.07476795 0.00781250 -0.76815462 -0.08869865 -0.04486077 0.00781250 -0.76815462 -0.08869865 -0.01495359 0.00781250 -0.76815462 0.26609597 -0.10467514 0.00781250 -0.76815462 0.26609597 -0.07476795 0.00781250 -0.76815462 0.26609597 -0.04486077 0.00781250 -0.76815462 0.26609597 -0.01495359 0.00781250 -0.76815462 0.62089056 -0.10467514 0.00781250 -0.76815462 0.62089056 -0.07476795 0.00781250 -0.76815462 0.62089056 -0.04486077 0.00781250 -0.76815462 0.62089056 -0.01495359 0.00781250 -0.76815462 0.97568518 -0.10467514 0.00781250 -0.76815462 0.97568518 -0.07476795 0.00781250 -0.76815462 0.97568518 -0.04486077 0.00781250 -0.76815462 0.97568518 -0.01495359 0.00781250 -0.76815462 1.33047974 -0.10467514 0.00781250 -0.76815462 1.33047974 -0.07476795 0.00781250 -0.76815462 1.33047974 -0.04486077 0.00781250 -0.76815462 1.33047974 -0.01495359 0.00781250 -0.76815462 -1.15308248 0.61309722 0.00781250 -0.76815462 -1.15308248 0.64300440 0.00781250 -0.76815462 -1.15308248 0.67291158 0.00781250 -0.76815462 -1.15308248 0.70281876 0.00781250 -0.46089280 -0.97568518 -0.10467514 0.00781250 -0.46089280 -0.97568518 -0.07476795 0.00781250 -0.46089280 -0.97568518 -0.04486077 0.00781250 -0.46089280 -0.97568518 -0.01495359 0.00781250 -0.46089280 -0.62089056 -0.10467514 0.00781250 -0.46089280 -0.62089056 -0.07476795 0.00781250 -0.46089280 -0.62089056 -0.04486077 0.00781250 -0.46089280 -0.62089056 -0.01495359 0.00781250 -0.46089280 -0.26609597 -0.10467514 0.00781250 -0.46089280 -0.26609597 -0.07476795 0.00781250 -0.46089280 -0.26609597 -0.04486077 0.00781250 -0.46089280 -0.26609597 -0.01495359 0.00781250 -0.46089280 0.08869865 -0.10467514 0.00781250 -0.46089280 0.08869865 -0.07476795 0.00781250 -0.46089280 0.08869865 -0.04486077 0.00781250 -0.46089280 0.08869865 -0.01495359 0.00781250 -0.46089280 0.44349325 -0.10467514 0.00781250 -0.46089280 0.44349325 -0.07476795 0.00781250 -0.46089280 0.44349325 -0.04486077 0.00781250 -0.46089280 0.44349325 -0.01495359 0.00781250 -0.46089280 0.79828787 -0.10467514 0.00781250 -0.46089280 0.79828787 -0.07476795 0.00781250 -0.46089280 0.79828787 -0.04486077 0.00781250 -0.46089280 0.79828787 -0.01495359 0.00781250 -0.46089280 1.15308249 -0.10467514 0.00781250 -0.46089280 1.15308249 -0.07476795 0.00781250 -0.46089280 1.15308249 -0.04486077 0.00781250 -0.46089280 1.15308249 -0.01495359 0.00781250 -0.46089280 -1.33047973 0.61309722 0.00781250 -0.46089280 -1.33047973 0.64300440 0.00781250 -0.46089280 -1.33047973 0.67291158 0.00781250 -0.46089280 -1.33047973 0.70281876 0.00781250 -0.15363093 -1.15308249 -0.10467514 0.00781250 -0.15363093 -1.15308249 -0.07476795 0.00781250 -0.15363093 -1.15308249 -0.04486077 0.00781250 -0.15363093 -1.15308249 -0.01495359 0.00781250 -0.15363093 -0.79828787 -0.10467514 0.00781250 -0.15363093 -0.79828787 -0.07476795 0.00781250 -0.15363093 -0.79828787 -0.04486077 0.00781250 -0.15363093 -0.79828787 -0.01495359 0.00781250 -0.15363093 -0.44349325 -0.10467514 0.00781250 -0.15363093 -0.44349325 -0.07476795 0.00781250 -0.15363093 -0.44349325 -0.04486077 0.00781250 -0.15363093 -0.44349325 -0.01495359 0.00781250 -0.15363093 -0.08869865 -0.10467514 0.00781250 -0.15363093 -0.08869865 -0.07476795 0.00781250 -0.15363093 -0.08869865 -0.04486077 0.00781250 -0.15363093 -0.08869865 -0.01495359 0.00781250 -0.15363093 0.26609597 -0.10467514 0.00781250 -0.15363093 0.26609597 -0.07476795 0.00781250 -0.15363093 0.26609597 -0.04486077 0.00781250 -0.15363093 0.26609597 -0.01495359 0.00781250 -0.15363093 0.62089056 -0.10467514 0.00781250 -0.15363093 0.62089056 -0.07476795 0.00781250 -0.15363093 0.62089056 -0.04486077 0.00781250 -0.15363093 0.62089056 -0.01495359 0.00781250 -0.15363093 0.97568518 -0.10467514 0.00781250 -0.15363093 0.97568518 -0.07476795 0.00781250 -0.15363093 0.97568518 -0.04486077 0.00781250 -0.15363093 0.97568518 -0.01495359 0.00781250 -0.15363093 1.33047974 -0.10467514 0.00781250 -0.15363093 1.33047974 -0.07476795 0.00781250 -0.15363093 1.33047974 -0.04486077 0.00781250 -0.15363093 1.33047974 -0.01495359 0.00781250
The Fireball.sample file from which fireball.in will be created looks like this:
&OPTION basisfile = Cu_bulk.bas lvsfile = Cu_bulk.lvs kptpreference = Cu_bulk.kpts nstepf = 1 rescal = AAA &END
And finally the running script named eg. fireball.sh is here (please note, that in you directory there has to be a file called runTG.com, which obtains path to the fireball.k.x executable, or the executable itselves):
alat="3.21 3.22 3.23 3.24 3.25 3.26 3.27 3.28 3.29 3.30 3.31 3.32 3.33 3.34 3.35 3.36 3.37 3.38 3.39" for i in $alat; do echo $i mkdir $i lattice="`echo $i`"; sed "s/AAA/$lattice/g" fireball.sample > fireball.in; ./runTG.com > a-$i.out en=`grep "etot/atom" a-$i.out | cut -b50-65` echo $i $en >> energy.dat mv CHARGES $i/ mv a-$i.out $i/ done
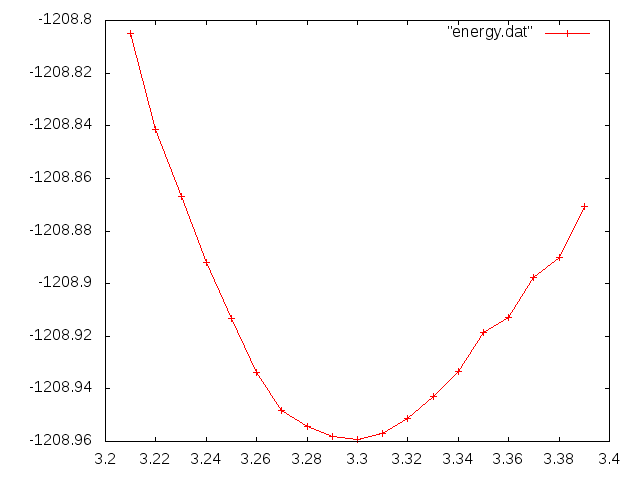
By simple plotting with gnuplot:
plot "energy.dat" w lp
, you'll obtain, that the optimized lattice constant is 3.3 Angstrom. (The experimental is 3.614 Angstrom.)