Table of Contents
Tutorial H-chain in PDF
There is simple presentation of smeagol usage presented on Smeagol Workshop in Hungary
(no much text explanatiobn included)
The input data files for this tutorial could be downloaded here
Wiki Tutorial on H-chain
Intro
Smeagol computation is split to 2 parts and 4 independent runs
- LEADs computation
- Fireball SCF (converge equlibrium density of LEADs)
- Export LEADs files
- SYSTEM computation
- Fireball SCF (converge equlibrium density of molecule with leads)
- Smeagol computation (Get current, conductivity, transmission spectra)
Currently there are 3 versions of smeagol implementation
- non-SCF smeagol - there you use equlibrium density from Fireball. It's much faster than nonequlibrium SCF-loop in smeagol. Currently this is the only version which looks to work fine.
- SCF with Kohn-Sham grid - This should be almost identical to siesta implementation of smeagol. Currently it looks working in principle but there are problems with discontinuity on bonundary of leads.
- McWeda smeagol - There is a problem with tranformation of overlap matrix, so consider it as non-working
As simplest example I will show computation of hydrogen molecule in between hydrogen leads
LEADS computation
Let's use this lead geometry
answer.bas
6 1 3.000000 3.000000 1.000000 1 3.000000 3.000000 2.000000 1 3.000000 3.000000 3.000000 1 3.000000 3.000000 4.000000 1 3.000000 3.000000 5.000000 1 3.000000 3.000000 6.000000
cel.lvs
20.000000000 0.000000000 0.000000000
0.000000000 20.000000000 0.000000000
0.000000000 0.000000000 6.000000000
because you have to converge density of lead is infinite system you should use good sampling in z-direction (NOTE: Smeagol always expect current to flow in z-direction !!!)
input.kpts
100 0.0000000000 0.0000000000 -0.5183627873 0.0100000000 0.0000000000 0.0000000000 -0.5078908118 0.0100000000 0.0000000000 0.0000000000 -0.4974188363 0.0100000000 0.0000000000 0.0000000000 -0.4869468608 0.0100000000 0.0000000000 0.0000000000 -0.4764748853 0.0100000000 0.0000000000 0.0000000000 -0.4660029098 0.0100000000 0.0000000000 0.0000000000 -0.4555309343 0.0100000000 0.0000000000 0.0000000000 -0.4450589588 0.0100000000 0.0000000000 0.0000000000 -0.4345869833 0.0100000000 0.0000000000 0.0000000000 -0.4241150078 0.0100000000 0.0000000000 0.0000000000 -0.4136430323 0.0100000000 0.0000000000 0.0000000000 -0.4031710568 0.0100000000 0.0000000000 0.0000000000 -0.3926990813 0.0100000000 0.0000000000 0.0000000000 -0.3822271058 0.0100000000 0.0000000000 0.0000000000 -0.3717551303 0.0100000000 0.0000000000 0.0000000000 -0.3612831548 0.0100000000 0.0000000000 0.0000000000 -0.3508111793 0.0100000000 0.0000000000 0.0000000000 -0.3403392038 0.0100000000 0.0000000000 0.0000000000 -0.3298672283 0.0100000000 0.0000000000 0.0000000000 -0.3193952528 0.0100000000 0.0000000000 0.0000000000 -0.3089232773 0.0100000000 0.0000000000 0.0000000000 -0.2984513018 0.0100000000 0.0000000000 0.0000000000 -0.2879793263 0.0100000000 0.0000000000 0.0000000000 -0.2775073507 0.0100000000 0.0000000000 0.0000000000 -0.2670353753 0.0100000000 0.0000000000 0.0000000000 -0.2565633998 0.0100000000 0.0000000000 0.0000000000 -0.2460914243 0.0100000000 0.0000000000 0.0000000000 -0.2356194488 0.0100000000 0.0000000000 0.0000000000 -0.2251474733 0.0100000000 0.0000000000 0.0000000000 -0.2146754978 0.0100000000 0.0000000000 0.0000000000 -0.2042035223 0.0100000000 0.0000000000 0.0000000000 -0.1937315468 0.0100000000 0.0000000000 0.0000000000 -0.1832595713 0.0100000000 0.0000000000 0.0000000000 -0.1727875958 0.0100000000 0.0000000000 0.0000000000 -0.1623156203 0.0100000000 0.0000000000 0.0000000000 -0.1518436448 0.0100000000 0.0000000000 0.0000000000 -0.1413716693 0.0100000000 0.0000000000 0.0000000000 -0.1308996937 0.0100000000 0.0000000000 0.0000000000 -0.1204277182 0.0100000000 0.0000000000 0.0000000000 -0.1099557427 0.0100000000 0.0000000000 0.0000000000 -0.0994837672 0.0100000000 0.0000000000 0.0000000000 -0.0890117917 0.0100000000 0.0000000000 0.0000000000 -0.0785398163 0.0100000000 0.0000000000 0.0000000000 -0.0680678407 0.0100000000 0.0000000000 0.0000000000 -0.0575958652 0.0100000000 0.0000000000 0.0000000000 -0.0471238897 0.0100000000 0.0000000000 0.0000000000 -0.0366519142 0.0100000000 0.0000000000 0.0000000000 -0.0261799387 0.0100000000 0.0000000000 0.0000000000 -0.0157079633 0.0100000000 0.0000000000 0.0000000000 -0.0052359878 0.0100000000 0.0000000000 0.0000000000 0.0052359878 0.0100000000 0.0000000000 0.0000000000 0.0157079633 0.0100000000 0.0000000000 0.0000000000 0.0261799388 0.0100000000 0.0000000000 0.0000000000 0.0366519143 0.0100000000 0.0000000000 0.0000000000 0.0471238898 0.0100000000 0.0000000000 0.0000000000 0.0575958653 0.0100000000 0.0000000000 0.0000000000 0.0680678408 0.0100000000 0.0000000000 0.0000000000 0.0785398163 0.0100000000 0.0000000000 0.0000000000 0.0890117917 0.0100000000 0.0000000000 0.0000000000 0.0994837672 0.0100000000 0.0000000000 0.0000000000 0.1099557427 0.0100000000 0.0000000000 0.0000000000 0.1204277182 0.0100000000 0.0000000000 0.0000000000 0.1308996937 0.0100000000 0.0000000000 0.0000000000 0.1413716692 0.0100000000 0.0000000000 0.0000000000 0.1518436448 0.0100000000 0.0000000000 0.0000000000 0.1623156203 0.0100000000 0.0000000000 0.0000000000 0.1727875958 0.0100000000 0.0000000000 0.0000000000 0.1832595713 0.0100000000 0.0000000000 0.0000000000 0.1937315468 0.0100000000 0.0000000000 0.0000000000 0.2042035223 0.0100000000 0.0000000000 0.0000000000 0.2146754977 0.0100000000 0.0000000000 0.0000000000 0.2251474732 0.0100000000 0.0000000000 0.0000000000 0.2356194487 0.0100000000 0.0000000000 0.0000000000 0.2460914242 0.0100000000 0.0000000000 0.0000000000 0.2565633997 0.0100000000 0.0000000000 0.0000000000 0.2670353753 0.0100000000 0.0000000000 0.0000000000 0.2775073508 0.0100000000 0.0000000000 0.0000000000 0.2879793263 0.0100000000 0.0000000000 0.0000000000 0.2984513018 0.0100000000 0.0000000000 0.0000000000 0.3089232773 0.0100000000 0.0000000000 0.0000000000 0.3193952528 0.0100000000 0.0000000000 0.0000000000 0.3298672283 0.0100000000 0.0000000000 0.0000000000 0.3403392038 0.0100000000 0.0000000000 0.0000000000 0.3508111793 0.0100000000 0.0000000000 0.0000000000 0.3612831548 0.0100000000 0.0000000000 0.0000000000 0.3717551303 0.0100000000 0.0000000000 0.0000000000 0.3822271058 0.0100000000 0.0000000000 0.0000000000 0.3926990813 0.0100000000 0.0000000000 0.0000000000 0.4031710568 0.0100000000 0.0000000000 0.0000000000 0.4136430323 0.0100000000 0.0000000000 0.0000000000 0.4241150078 0.0100000000 0.0000000000 0.0000000000 0.4345869833 0.0100000000 0.0000000000 0.0000000000 0.4450589588 0.0100000000 0.0000000000 0.0000000000 0.4555309343 0.0100000000 0.0000000000 0.0000000000 0.4660029098 0.0100000000 0.0000000000 0.0000000000 0.4764748853 0.0100000000 0.0000000000 0.0000000000 0.4869468607 0.0100000000 0.0000000000 0.0000000000 0.4974188362 0.0100000000 0.0000000000 0.0000000000 0.5078908117 0.0100000000 0.0000000000 0.0000000000 0.5183627872 0.0100000000
first you have to run SCF calculation to converge charges, use this input file Fireball.in (scf)
&OPTION basisfile = answer.bas lvsfile = cel.lvs icluster = 0 nstepf = 1 sigmatol = 0.0000000001 max_scf_iterations = 100 iqout = 1 ismeagol = 0 ifixcharge = 0 &END &OUTPUT iwrtHSrho = 0 iwrteigen = 1 iwrtdos = 0 &END
then fix charges, activate iwrtHSrho = 1 and run computation again to export LEADs density
&OPTION basisfile = answer.bas lvsfile = cel.lvs icluster = 0 nstepf = 1 sigmatol = 0.0000000001 max_scf_iterations = 100 dt = 0.5 iqout = 1 ismeagol = 0 ifixcharge = 1 &END &OUTPUT iwrtHSrho = 1 iwrteigen = 0 iwrtdos = 0 &END
to do this you have to specify k-point sampling of the SYSTEM you want to use the LEADs in. This kpoints file is called MOLECULE.kpts. In our case it has just gamma point, however in general it could have any sampling in x and y direction. (never in z!!).
MOLECULE.kpts
1 0.0000000000 0.0000000000 0.0000000000 1.0000000000
after the computation you get file called ELECTRODE where k-space representations of Hamiltonian, Density matrix and overplap matrix are stored.
SYSTEM computation
Your system should contain the geometry of leads in it's geometry description, it should also contain some region where charge redistribution is screened in order to smoothly align with LEADs, because LEADs itselfs represent infinite bulk and their chrage distribution is fixed during the computation. Order of atoms MUST be exactly the same as in LEADs calculation and left lead must be at the begining, and right lead must be at the end of file. For example:
answer.bas
24 1 3.000000000 3.000000000 1.000000000 #start LEAD.left 1 3.000000000 3.000000000 2.000000000 1 3.000000000 3.000000000 3.000000000 1 3.000000000 3.000000000 4.000000000 1 3.000000000 3.000000000 5.000000000 1 3.000000000 3.000000000 6.000000000 # end LEAD.left 1 3.000000000 3.000000000 7.000000000 # screening region (left) 1 3.000000000 3.000000000 8.000000000 1 3.000000000 3.000000000 9.000000000 1 3.000000000 3.000000000 10.000000000 1 3.000000000 3.000000000 11.000000000 1 3.000000000 3.000000000 13.000000000 # molecule itselfs 1 3.000000000 3.000000000 14.000000000 1 3.000000000 3.000000000 16.000000000 # screening region (right) 1 3.000000000 3.000000000 17.000000000 1 3.000000000 3.000000000 18.000000000 1 3.000000000 3.000000000 19.000000000 1 3.000000000 3.000000000 20.000000000 1 3.000000000 3.000000000 21.000000000 #start LEAD.right 1 3.000000000 3.000000000 22.000000000 1 3.000000000 3.000000000 23.000000000 1 3.000000000 3.000000000 24.000000000 1 3.000000000 3.000000000 25.000000000 1 3.000000000 3.000000000 26.000000000 # end LEAD.left
cel.lvs
20.000000000 0.000000000 0.000000000
0.000000000 20.000000000 0.000000000
0.000000000 0.000000000 26.000000000
Your kpoint sampling MUST be the same as MOLECULE.kpts in LEADs calculation. Otherweis ELECTRODE files are incompatible. input.kpts
1 0.0000000000 0.0000000000 0.0000000000 1.0000000000
first you should run SCF run with fireball (with smeagol off), it's much faster than doing smeagol directly from neutral atom charges. Also, currently smeagol SCF implementation in fireball is not perfect.
fireball.in (scf)
&OPTION basisfile = answer.bas lvsfile = cel.lvs icluster = 0 nstepf = 1 sigmatol = 0.0000000001 max_scf_iterations = 100 iqout = 1 ismeagol=0 &END &OUTPUT iwrtHSrho = 0 &END
After convergence of charges, you need to copy ELECTRODE files from LEADs calculation. Rename it ELECTRODE.left, and ELECTRODE.right. You can use different leads on left and right. Both left and right lead should have the same order (top to down in z-direction) it means right lead SHOULD NOT be in reverse order (mirror).
Then you can run the smeagol calculation. To do this set ismeagol = 1 in fireball.in.
fireball.in
&OPTION basisfile = answer.bas lvsfile = cel.lvs icluster = 0 nstepf = 1 sigmatol = 0.000001 max_scf_iterations = 199 dt = 0.5 iqout = 1 ismeagol=1 tempfe=300 &END &OUTPUT iwrtHSrho = 0 &END
You should also specify smeagol parameters in smeagol.optional
500 NEnergR 90 NEnergIC 20 NEnergIL 10 NPoles 0.001 Delta -45.0 EnergLB 1 NSlices T TrCoeff 1000 NeneT -20.0 TEnergI 30.0 TEnergF -4.3217 Fermi_level 0.1 V_Bias 12.0 r_left 12.5 r_right 1 useLeads? 4.50 r_start_fithop 0.25 r_scale_fithop
useLeads shoudl be always set to 1 otherweise the Transition spectrum can not be computed ( 0 is just for debug purpose)
most of the parameters should be used default. Only what you should care about EnergLB - should be set reasonably lower than lowest energy in molecular spectrum. Be ware that in case of non-equlibrium selfconsistency computation the levels energy could change considerably. If some level become lower than EnergLB the selfconsistency would not conserve charges makes it imposible to converge. TrCoeff (T for true, F for False) specify if Transmission spectrum should be ploted (set T most of the time) NeneT is number of points in transmisison spectrum and TEnergI and TEnergF minimum an maximum of energy interval.
In case of non-selfconsistent computation (which we discribe here) you should set also Fermi_level to fermilevel of LEADs (from your LEADs calculation). If you are interested in current at nonzero voltage, set VBias and r_left, r_right to specify potential ramp which is addet to hamiltonian. Potential ramp is
- V = -VBias/2 for -infinity< r <r_left
- V = -VBias/2 + (r - r_left)/(r_right - r_left) for r_right < r <r_left
- V = +VBias/2 for r_right < r < +infinity
At Last, there is possibylity to fit extended basis-functions at apex region in order to mimic realistic decay in vacuum for non-contact tuneling current computation in bigger distance.
To do this you have to provide interaction.optional and files contanting overlap interaction_S_X_Y.optional and interaction hamiltonian interaction_H_X_Y.optional for pairs of elements X,Y. for Example you should provide interaction_S_79_79.optional nad interaction_H_79_79.optional for interaction of two gold electrodes.
The method of is automatically switched on if interaction.optional is provided, and of if it is not avaible in the directory.
Than you should set up transition betwen original basisfunctions and the new one in smeagol.optional. r_start_fithop define distace where the hopping integral start to change from standard to extended. r_scale_fithop define width of the intermediate region. r_scale_fithop=0.0 means stepwise change.
After do computation otuptfiles smeagol.CUR and smeagol.TRC are generated. .CUR for current and .TRC for transmission coefficient.
Format of output files is as flows
# V = 0.0071 k-points: 1 -0.1567831D+02 0.2257147D-30 0.2257147D-30 0 -0.1562826D+02 0.2227090D-30 0.2227090D-30 0 -0.1557821D+02 0.2197281D-30 0.2197281D-30 0 -0.1552816D+02 0.2167719D-30 0.2167719D-30 0 -0.1547811D+02 0.2138402D-30 0.2138402D-30 0 -0.1542806D+02 0.2109329D-30 0.2109329D-30 0 -0.1537801D+02 0.2080497D-30 0.2080497D-30 0
first column is Energy in [eV], second and third are Transmission coefficient for spin Up and Down. Currently computation is not spin resolved, and both colums are identical.
in .CUR is current (second column) and for given voltage (first column)
0.70561919D-02 0.19849858D-06
at the moment curent is computed always for one voltage. In future there should be increasing voltage from zero (equlibrium) gradually to maximum voltage. This would be effective for convergence of nonequlibrium density.
Scripted tutorial on BiThioBenzene on gold 100
After finishing H-chain tutorial (wich computational time ~20 second is short enought for learning purpose ), you can approach to much more computationaly demanding task of computation of transmission coefficient for molecule between gold electrodes. The computationl time of this example would be arround 1 day, but can by shotened by setting shorter and less densly sampled energy region in smeagol optional by
1000 NeneT -20.0 TEnergI 30.0 TEnergF
All the input files and a script to successively execute all the neccesary steps is provided in this .zip package.
In order to run this tutorial it is necessary to make a link of Fdata in both 'Au100-LEADS-1k' and 'AuBTB-1k' directory.
than you have just tu run the script run_all. The script is suposed to be submitted to the PBS front system, form the directory and use variable $PBS_O_WORKDIR for correct function.
If you are going to run it interactively outside the PBS front system, remove the line
cd $PBS_O_WORKDIR
form the script
We can comment successive compuatational steps in script now.
#! /bin/bash LEADSdir="Au100-LEADS-1k" SYSTEMdir="AuBTB-1k" cd $PBS_O_WORKDIR # ================= 1 ====================== # # Start LEADS computation cd $LEADSdir # remove old files rm NEIGHBORS rm NEIGHBORS_PP rm ELECTRODE* rm fort.* rm core.* rm ERR* rm OUT* rm CHARGES* # Compute self consistent equlibirum CHARGE distribution in LEADS cp fireball.in-scf fireball.in ../fireball >OUTscf 2>ERRscf # Export ELECTRODE files cp fireball.in-leads fireball.in ../fireball >OUTleads 2>ERRleads # end LEADS computation cd .. # ================= 2 ====================== # # Copy files from LEADS to SYSTEM cp -f $LEADSdir/ELECTRODE $SYSTEMdir/ELECTRODE.left cp -f $LEADSdir/ELECTRODE $SYSTEMdir/ELECTRODE.right cp -f $LEADSdir/CHARGES $SYSTEMdir/CHARGES.left cp -f $LEADSdir/CHARGES $SYSTEMdir/CHARGES.right cp -f $LEADSdir/MOLECULE.kpts $SYSTEMdir/input.kpts # set LEADS fermilevel for smeagl fermi=`grep Fermi $LEADSdir/OUTscf | tail -1 | cut -b 17-` sed "s/AAA/$fermi/g" $SYSTEMdir/smeagol.optional-0 > $SYSTEMdir/smeagol.optional # ================= 3 ====================== # # Start SYSTEM computation cd $SYSTEMdir # remove old files rm NEIGHBORS rm NEIGHBORS_PP rm fort.* rm core.* rm ERR* rm OUT* rm CHARGES rm smeagol.TRC # Compute self consistent equlibirum CHARGE distribution of SYSTEM cp fireball.in-scf fireball.in ../fireball >OUTscf 2>ERRscf # Smeagol computation cp fireball.in-smeagol fireball.in ../fireball >OUTsmeagol 2>ERRsmeagol
We recommand you to follow the script order line by line, watching on the description of manual succesive executing of each steps which is described in H-chain tutorial section above. We also recommand you to examine imput files like
Au100-LEADS-1k/fireball.in-scf Au100-LEADS-1k/fireball.in-leads Au100-LEADS-1k/answer.bas AuBTB-1k/fireball.in-scf AuBTB-1k/fireball.in-smeagol AuBTB-1k/smeagol.optional AuBTB-1k/answer.bas
which determine the computation.
We provide this script as a template for furter modification for specific computations you are going to do.
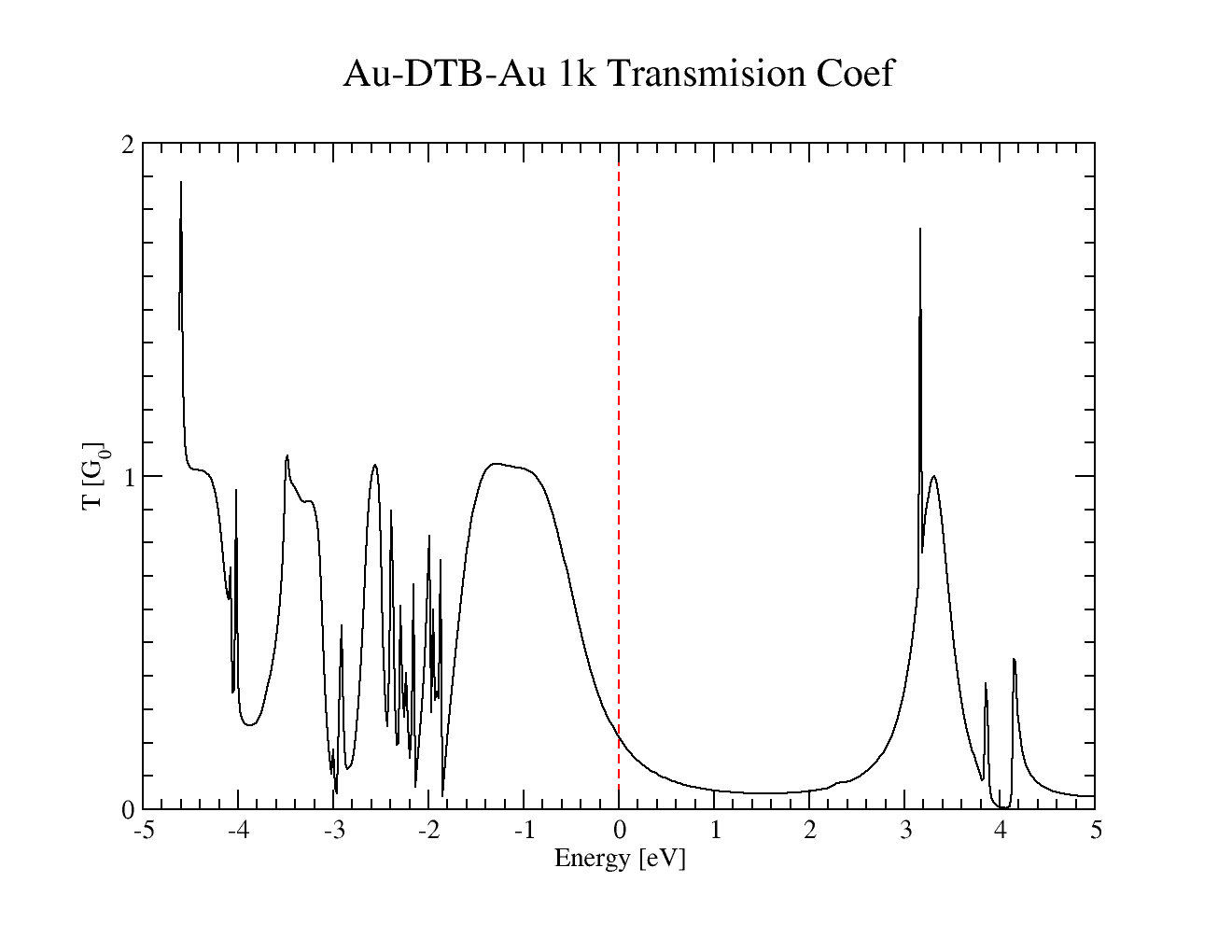
finally, after the computation is successfully finished, you should plot the outputfile with transmission coefficient
AuBTB-1k/smeagol.TRC
the plot for our basis set looks like this, but it can differ slightly with basiset
Transmussion coefficient for Dithiobenzene between gold 100 electrodes with 1 k-point. Basis used was H_s3.8 C_s4.0_p4.5_d5.40 S_s4.2_p4.7_d5.5 Au_s5.0_p5.6_d4.7