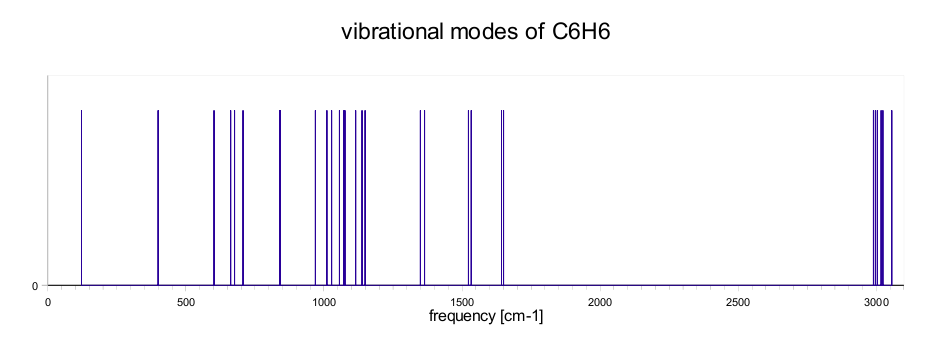
Vibrational modes
In this section we describe how to calculate the vibrational modes of molecules using dynamical matrix. Vibrational modes are obtained from eigenvalues of the dynamical matrix. The elements of the matrix are defined as second derivatives of energy with respect to geometry. The first derivatives with respect to geometry are forces.The second derivatives are obtained from the different of two forces and displacement. At first calculate optimization structure of the molecule and then move with atom in first direction and do SCF calculation, after that move with the same atom but in opposite direction and do again SCF calculation. Then evaluate the matrix element as different of this two forces devided by displacement. This procedure repeat for each direction and each atom of molecule. In order to calculate vibrational modes the dynamical matrix is mass weighted. Vibrational modes are calculated from eigenvalues of dynamical matrix.
Dynamical matrix calculation set the keywords idynmat in section &OPTION& in input file fireball in
fireball.in:
&OPTION basisfile = answer.bas icluster = 1 nstepf = 1 sigmatol = 0.00000001 max_scf_iterations = 100 dt = 0.5 iqout = 1 idynmat = 1 &END
Parameters of dynamical matrix calculation are set in file dyn.optional placed in work directory.
Calculation of all vibrational modes
dyn.optional:
0.03 !elementary displacement
1 1 1 !dimension vector
dyn.dat !output file
.false. !flag on when only some of atoms are moved, otherwise move all atoms
Calculation of vibrational modes only for selected atoms