Lattice parameter
In this section, we explain how to optimize the lattice parameter of Si bulk diamond structure. The diamond structure is characterized by two distinct atom per unit cell with relative coordinates (0 ,0 ,0) and (1/4, 1/4, 1/4). The lattice vector is defined in the relative coordinates alat*(1/4, 1/4, 1/4).
Detail description of the diamond structure can be found elsewhere.
There is an elegant way to rescale atomic coordinates, lattice vector and k-points by only one parameter rescal defined in the section &OPTIONS.
Our input atomic coordinates written in Si.bas look like:
2 14 0.00000 0.00000 0.00000 14 0.25000 0.25000 0.25000
the lattice vector is defined in Si.lvs file:
0.50000 0.00000 0.50000 0.00000 0.50000 0.50000 0.50000 0.50000 0.00000
and we use Oh symmetry group to generate a k-points set (file Si.kpts)
60 -2.74889350 -2.74889350 -2.74889350 0.00390625 -1.96349549 -1.96349549 -3.53429174 0.01171875 -1.17809725 -1.17809725 -4.31968975 0.01171875 -0.39269909 -0.39269909 -5.10508823 0.01171875 0.39269909 0.39269909 -5.89048624 0.01171875 1.17809725 1.17809725 5.89048608 0.01171875 1.96349549 1.96349549 5.10508807 0.01171875 -3.53429190 -3.53429190 -1.96349581 0.01171875 -2.74889350 -1.17809725 -2.74889350 0.01171875 -1.96349549 -0.39269909 -3.53429174 0.02343750 -1.17809725 0.39269909 -4.31968975 0.02343750 -0.39269909 1.17809725 -5.10508823 0.02343750 0.39269909 1.96349549 -5.89048624 0.02343750 -5.10508815 -3.53429190 -0.39269932 0.02343750 -4.31968991 -2.74889366 -1.17809733 0.02343750 -2.74889350 0.39269909 -2.74889350 0.01171875 -1.96349549 1.17809725 -3.53429174 0.02343750 -1.17809725 1.96349549 -4.31968975 0.02343750 -0.39269909 2.74889350 -5.10508823 0.02343750 -5.89048631 -2.74889366 0.39269916 0.02343750 -5.10508815 -1.96349565 -0.39269932 0.02343750 -2.74889350 1.96349549 -2.74889350 0.01171875 -1.96349549 2.74889350 -3.53429174 0.02343750 -1.17809725 3.53429174 -4.31968975 0.02343750 5.89048631 -1.96349565 1.17809717 0.02343750 -5.89048631 -1.17809717 0.39269916 0.02343750 -2.74889350 3.53429174 -2.74889350 0.01171875 4.31968991 -1.96349565 2.74889366 0.02343750 5.10508815 -1.17809717 1.96349565 0.02343750 3.53429190 -1.17809717 3.53429190 0.01171875 4.31968991 -0.39269916 2.74889366 0.02343750 3.53429190 0.39269932 3.53429190 0.01171875 -1.96349549 -1.96349549 -1.96349549 0.00390625 -1.17809725 -1.17809725 -2.74889350 0.01171875 -0.39269909 -0.39269909 -3.53429174 0.01171875 0.39269909 0.39269909 -4.31968975 0.01171875 1.17809725 1.17809725 -5.10508823 0.01171875 -4.31968991 -4.31968991 0.39269916 0.01171875 -1.96349549 -0.39269909 -1.96349549 0.01171875 -1.17809725 0.39269909 -2.74889350 0.02343750 -0.39269909 1.17809725 -3.53429174 0.02343750 0.39269909 1.96349549 -4.31968975 0.02343750 1.17809725 2.74889350 -5.10508823 0.02343750 -1.96349549 1.17809725 -1.96349549 0.01171875 -1.17809725 1.96349549 -2.74889350 0.02343750 -0.39269909 2.74889350 -3.53429174 0.02343750 0.39269909 3.53429174 -4.31968975 0.02343750 -1.96349549 2.74889350 -1.96349549 0.01171875 -1.17809725 3.53429174 -2.74889350 0.02343750 -1.96349549 4.31968975 -1.96349549 0.01171875 -1.17809725 -1.17809725 -1.17809725 0.00390625 -0.39269909 -0.39269909 -1.96349549 0.01171875 0.39269909 0.39269909 -2.74889350 0.01171875 1.17809725 1.17809725 -3.53429174 0.01171875 -1.17809725 0.39269909 -1.17809725 0.01171875 -0.39269909 1.17809725 -1.96349549 0.02343750 0.39269909 1.96349549 -2.74889350 0.02343750 -1.17809725 1.96349549 -1.17809725 0.01171875 -0.39269909 -0.39269909 -0.39269909 0.00390625 0.39269909 0.39269909 -1.17809725 0.01171875
to make the total energy calculation of several lattice parameters automatic, we use a script:
alat=" 5.0 5.1 5.2 5.3 5.4 5.5 5.6 5.7 5.8 5.9 6.0" for i in $alat; do echo $i mkdir $i lattice="`echo $i`"; sed "s/AAA/$lattice/g" fireball.sample > fireball.in; ./runTG.com > a-$i.out grep "etot/atom" a-$i.out | cut -b50-65 mv CHARGES $i/ mv a-$i.out $i/ done
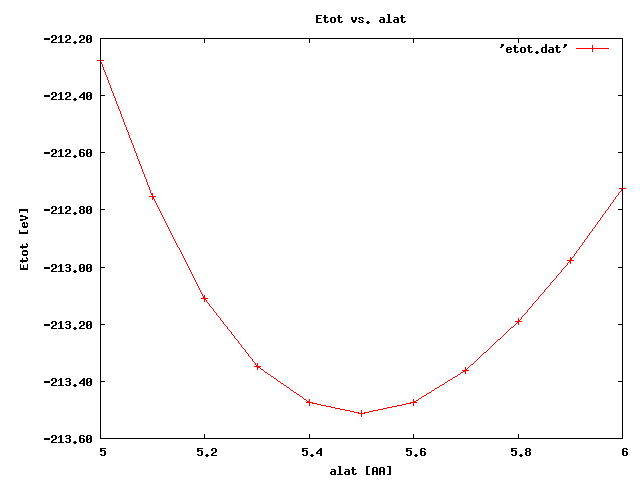
the script runs several tasks for defined lattice vectors, it creates a directory for each lattice parameter and it saves there CHARGES and an output file. It also dumps the energy per atom for a given lattice parameter.
A file fireball.sample has following syntax:
&OPTION basisfile = Si.bas lvsfile = Si.lvs kptpreference = Si.kpts nstepf = 1 rescal = AAA &END
Afterwards, we simply dump the total energy vs. lattice parameter into a file:
grep ETOT */*.out | cut -b1-3,34- > etot.dat
and we plot using gnuplot
gnuplot gnuplot> set title "Etot vs. alat" gnuplot> set xlabel "alat [Ang]" gnuplot> set ylabel "Etot [eV]" gnuplot> set format y "%8.2f" gnuplot> plot 'etot.dat' with linespoints
The bulk modulus
$ ls Fdata uno.bas uno.kpts uno.lvs vol.sh $ head uno.bas 2 14 0.000000 0.000000 0.000000 14 0.250000 0.250000 0.250000 $ head uno.kpts 32 -2.35619449 -2.35619449 -2.35619449 0.03125000 -3.92699081 -0.78539816 -0.78539816 0.03125000 -5.49778713 0.78539816 0.78539816 0.03125000 -7.06858347 2.35619449 2.35619449 0.03125000 -0.78539816 -3.92699081 -0.78539816 0.03125000 -2.35619449 -2.35619449 0.78539816 0.03125000 -3.92699081 -0.78539816 2.35619449 0.03125000 -5.49778713 0.78539816 3.92699081 0.03125000 0.78539816 -5.49778713 0.78539816 0.03125000 $ head uno.lvs 0.5000 0.5000 0.0000 0.5000 0.0000 0.5000 0.0000 0.5000 0.5000
The script to calculate the bulk modulus
$ cat vol.sh
#!/bin/bash
##----- Parametros de control (el parametro de red tiene que encontrase entre ini fin --------##
N=10
ini=5.0
fin=6.0
##----------Funcion analisis para dos atomos/celda-----------------------------------------##
function analisis {
ETOT=$(grep 'ETOT' salida.out|cut -d'=' -f2)
sigma=$(grep sigma salida.out | cut -d'=' -f16 | tail -1)
charge=$(head -2 uno.bas | tail -1 | tr -s ' ' | cut -d' ' -f2)' -> '$(head -2 CHARGES | tail -1)
charge=$charge' ;'$(head -3 uno.bas | tail -1 | tr -s ' ' | cut -d' ' -f2)' -> '
charge=$charge$(tail -1 CHARGES)
echo $rescal$'\t'$ETOT$'\t'$sigma$'\t'$charge$'\t'>>salida
}
##------------------------------------------------------------------------------------------##
function start {
rm -fr salida
for((i=0;i<=N;i++))
do
rescal=$(python -c "print '%.6f' % ($i*1.0*($fin-$ini)/$N+$ini)")
echo "&option
basisfile = uno.bas
lvsfile = uno.lvs
kptpreference = uno.kpts
rescal = $rescal
sigmatol = 0.000001
nstepf = 1
&end
&output
iwrtxyz = 1
&end" > fireball.in
../progs/fireball.x > salida.out
analisis
done
}
##----------------------------------1º start ----------------------------------------------##
start
##-----------------------------buscamos el minimo ------------------------------------------##
min=$(python -c "
x0 = []
y0 = []
for line in file(\"salida\"):
line = line.split()
x = line[0]
y = line[1]
x0.append(x)
y0.append(y)
j=0
for i in range(len(x0)):
if y0[j]< y0[i]:
j=i
print x0[j]")
cp salida borrar
rm -fr aux.py
##----------------------------------2º start -----------------------------------------------##
N=40
d=0.4
ini=$(python -c "print '%.6f' % (1.0*($min-$d))")
fin=$(python -c "print '%.6f' % (1.0*($min+$d))")
start
mv salida Vol.dat
##------------------------------------------------------------------------------------------##
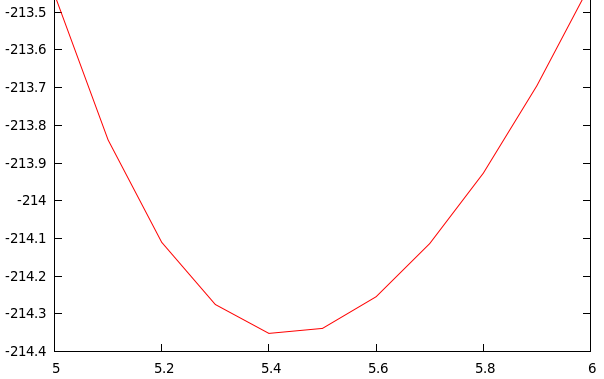
In the 1º start of the script “vol.sh” takes 10 points between 5-6:
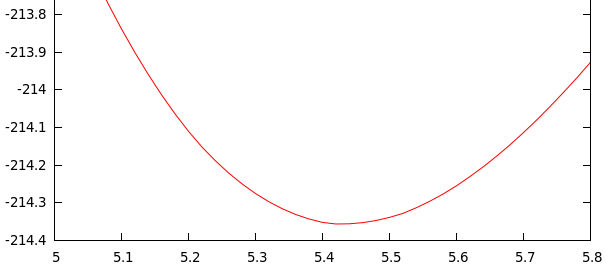
In the 2º start of the script “vol.sh” takes 40 points between the minimum of the 1º start
The scripts uses the parameter rescal in fireball.in and unitary positions and lattice vectors with 2 atoms/cell to obtain the kpts points we used xeo:
The bulk mudulus can be calculed also with xeo (Utilities→Bulk modulus), in the in line command:
$ xeo -Bulk Vol.dat zincblende E_min = -214.35239 eV a_min = 5.43719 angs Volumen = 40.18518 ang**3 Bulk modulus = 100.79399 GPa